![68Ga]ABY-028: an albumin-binding domain (ABD) protein-based imaging tracer for positron emission tomography (PET) studies of altered vascular permeability and predictions of albumin-drug conjugate transport | EJNMMI Research | Full Text 68Ga]ABY-028: an albumin-binding domain (ABD) protein-based imaging tracer for positron emission tomography (PET) studies of altered vascular permeability and predictions of albumin-drug conjugate transport | EJNMMI Research | Full Text](https://media.springernature.com/lw685/springer-static/image/art%3A10.1186%2Fs13550-020-00694-2/MediaObjects/13550_2020_694_Fig1_HTML.png)
68Ga]ABY-028: an albumin-binding domain (ABD) protein-based imaging tracer for positron emission tomography (PET) studies of altered vascular permeability and predictions of albumin-drug conjugate transport | EJNMMI Research | Full Text

In Silico Prediction of Compounds Binding to Human Plasma Proteins by QSAR Models - Sun - 2018 - ChemMedChem - Wiley Online Library
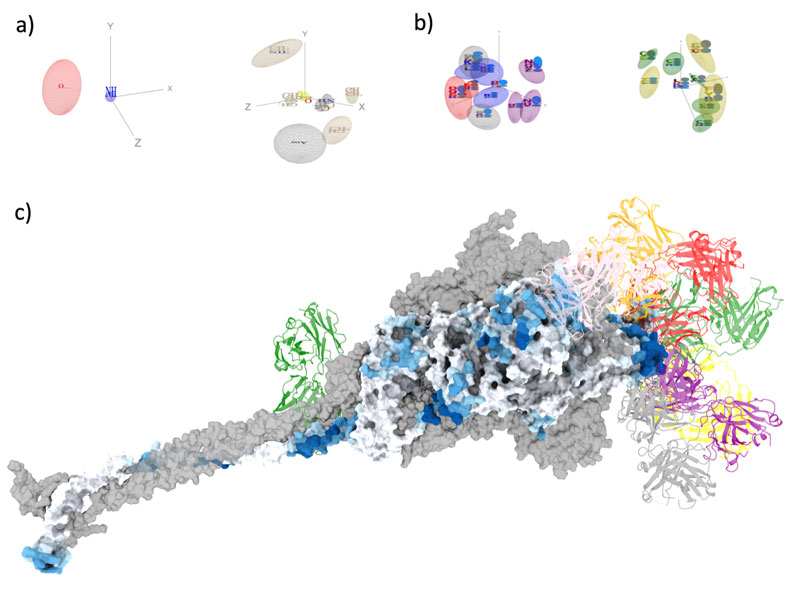
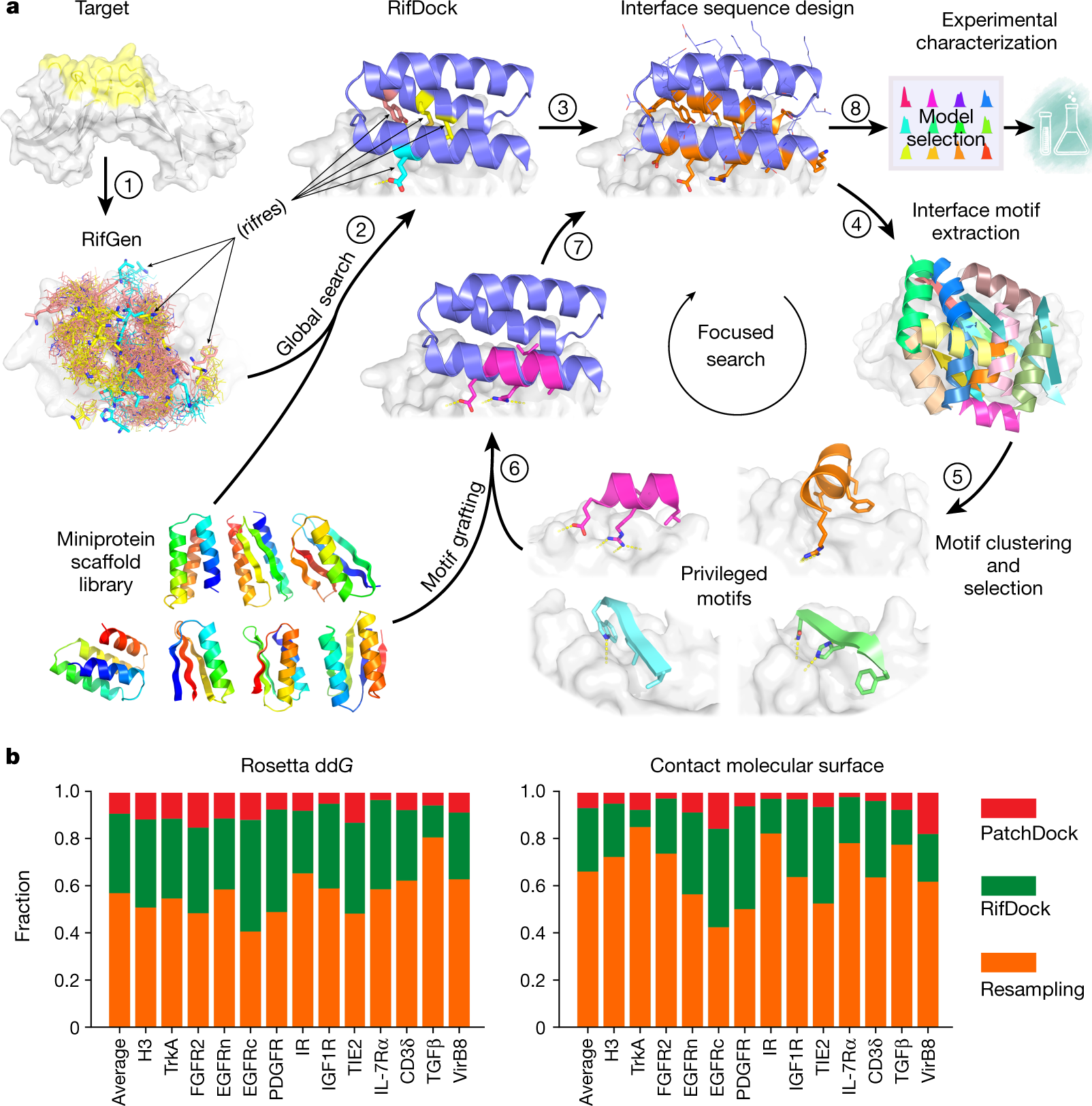
Mapping antibody epitopes onto protein structures with deep learning | Human Frontier Science Program
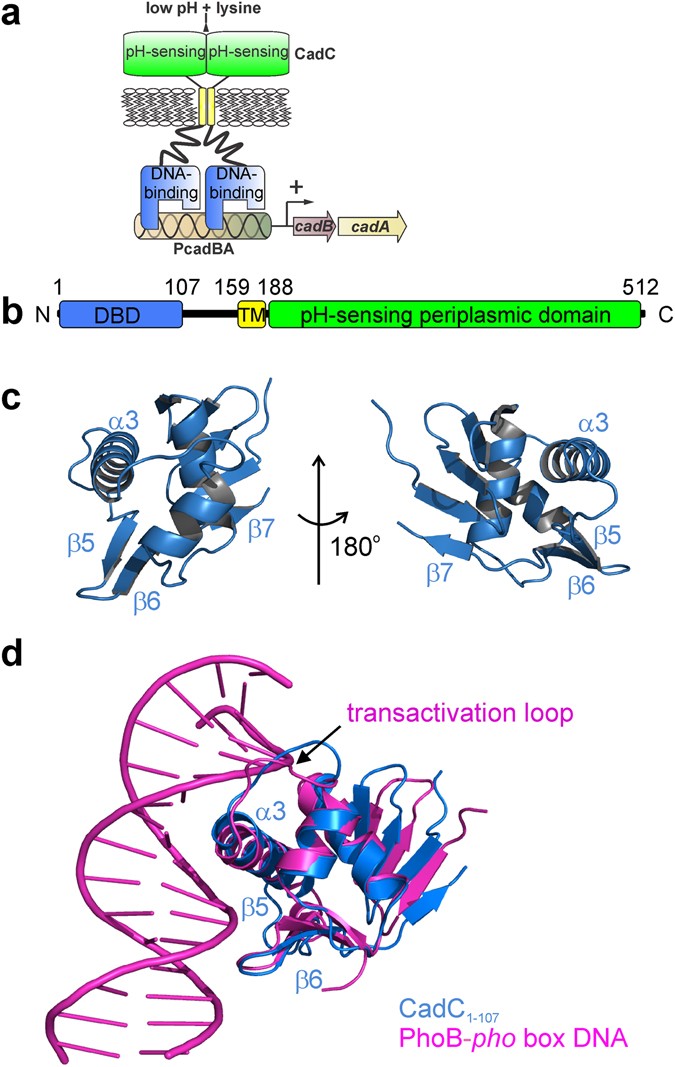
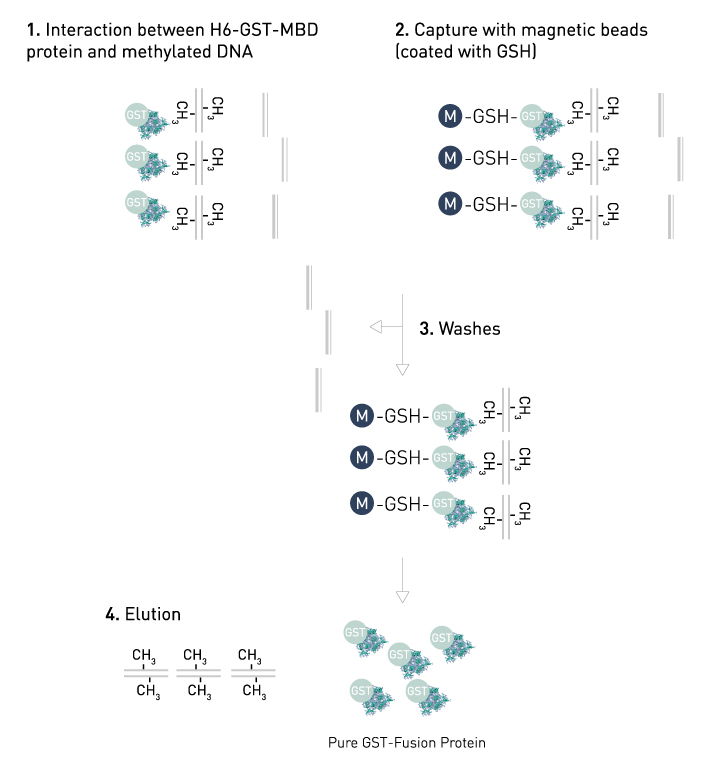
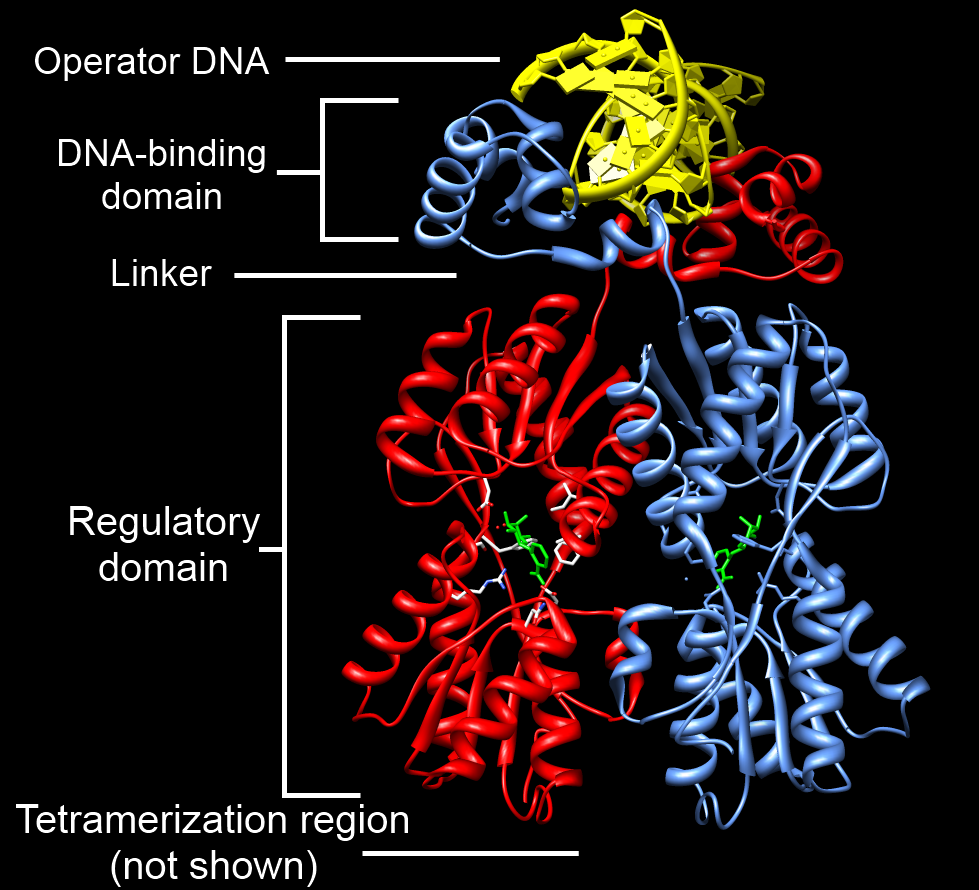
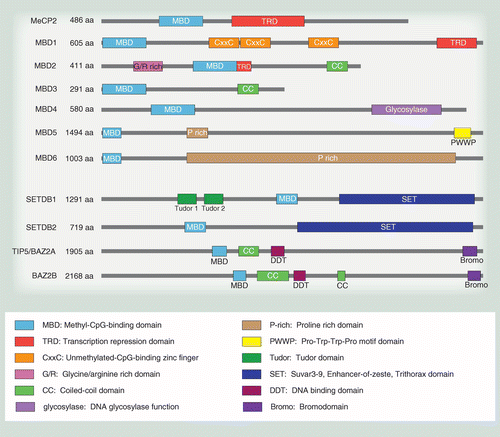
Structure-function analysis of the DNA-binding domain of a transmembrane transcriptional activator | Scientific Reports

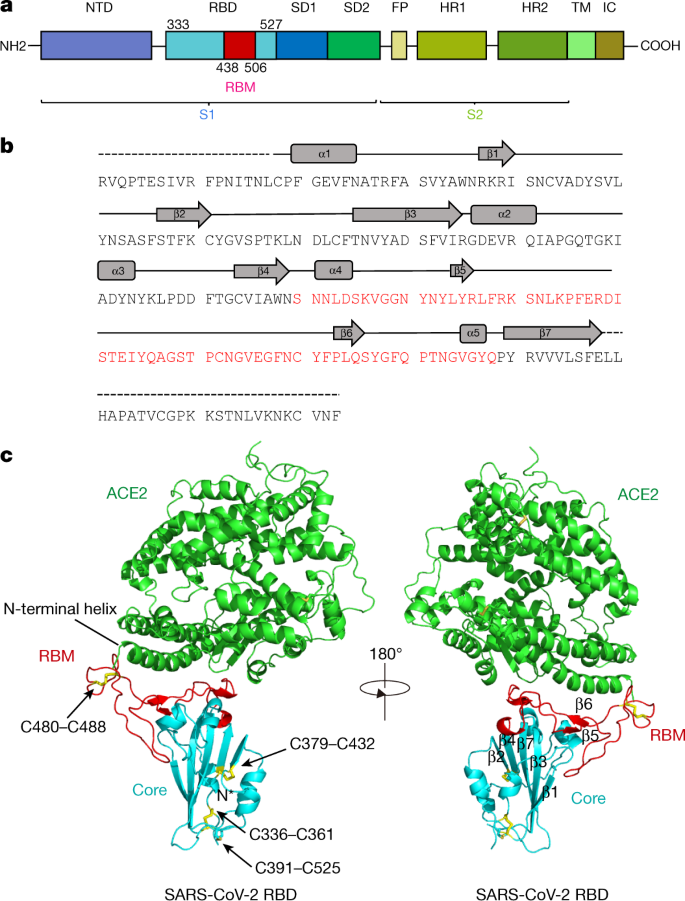
Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor | Nature Communications

Structural features of the protein kinase domain and targeted binding by small-molecule inhibitors - Journal of Biological Chemistry

Staphylococcal protein A inhibits complement activation by interfering with IgG hexamer formation | PNAS

A Promiscuous Lipid-Binding Protein Diversifies the Subcellular Sites of Toll-like Receptor Signal Transduction: Cell
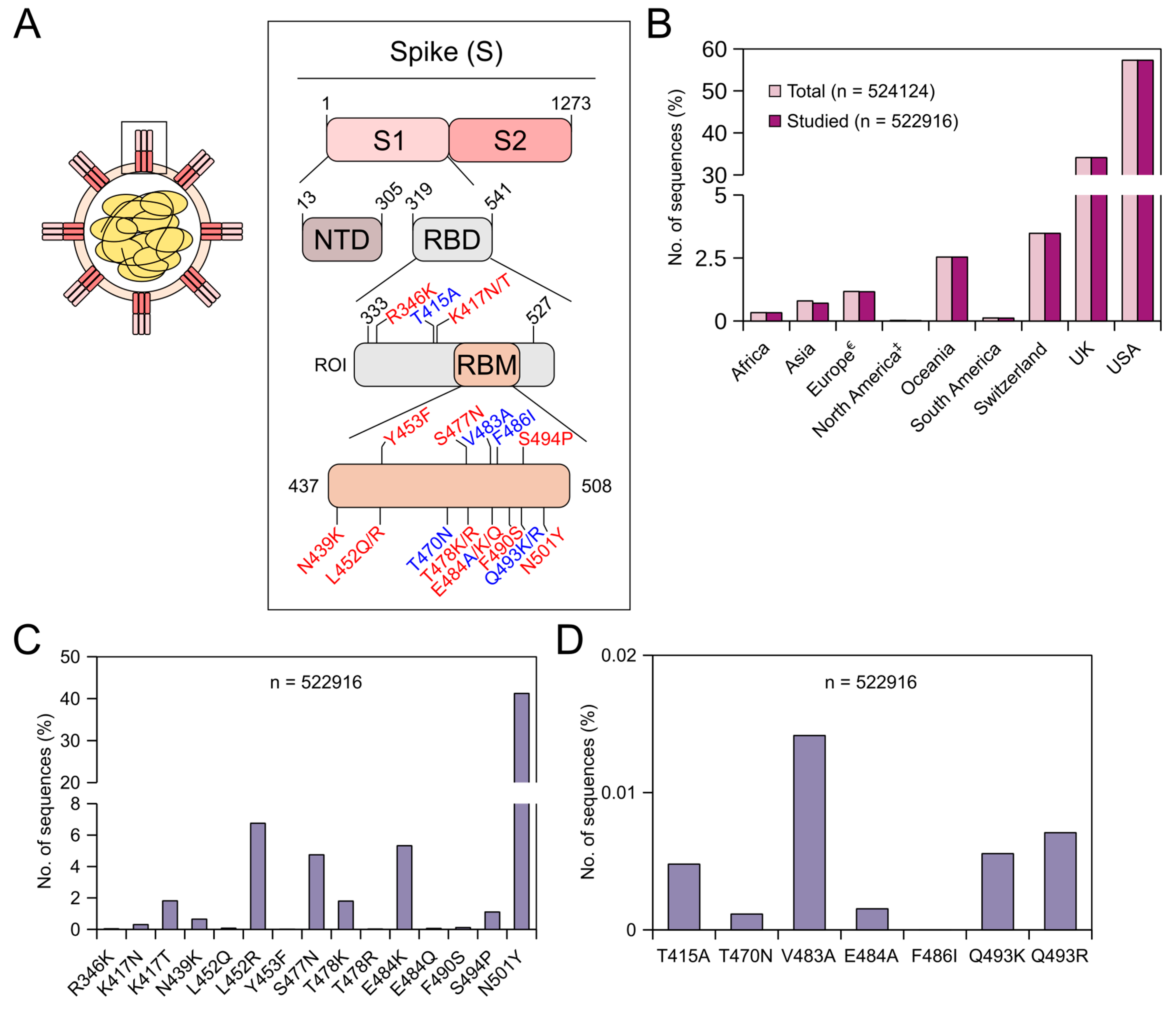
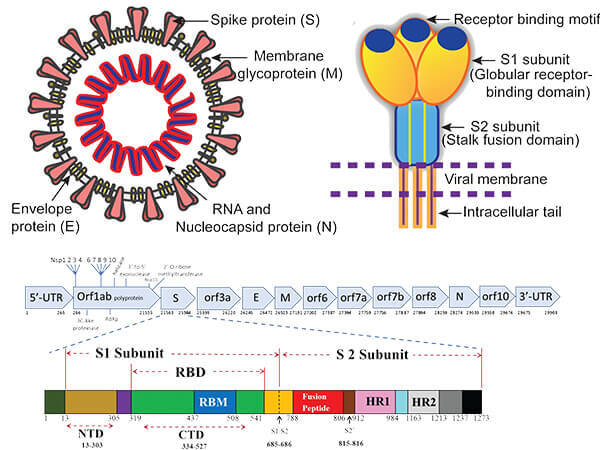
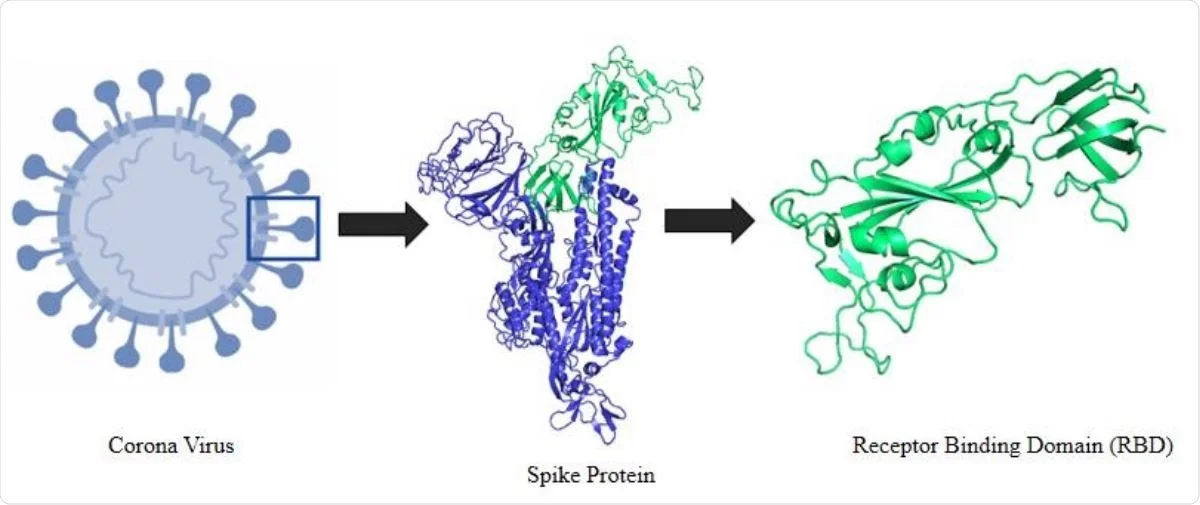
Viruses | Free Full-Text | Global Prevalence of Adaptive and Prolonged Infections' Mutations in the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein

Designing of peptide aptamer targeting the receptor-binding domain of spike protein of SARS-CoV-2: an in silico study | SpringerLink

Rapid Development of SARS-CoV-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates | ACS Nano